|
Genetic medicine and the application of rapid diagnostic techniques | Using IHGs for genetic diagnosis | SNPs and mutations | How is IHG technology relevant to SNP and mutation detection? | How does the IHG reagent work? | Examples: clinical PAGE minigels | What are the advantages of IHG technology? | How IHG technology compares | How can a single IHG reagent identify 2 or more individual SNPs and haplotypes? | Using IHGs for haplotyping | |
||||
Genetic medicine and the application of rapid diagnostic techniques |
||||
|
Genetic medicine embraces key disciplines such as: • Diagnosis of genetic disease • Determination of genetic risk factors • Forensic medicine • Pharmacogenetics – where the genetic configuration of an individual determines how that individual responds to a particular compound. This is of particular importance in cancer therapy, where a patient’s genetic configuration profoundly influences the success of a particular treatment, or treatment regime. |
||||
|
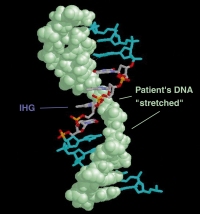
To take advantage of the progress made in these disciplines, there is a requirement for accurate, rapid and easy to use diagnostic methodology. IHG Pharmaco has developed a patented technology using induced heteroduplex generators (IHG) to screen for SNPs. The basic principle of the technology are shown in the illustration below, which shows a model of a gene with a mutation specific for an inherited genetic disease. One of the two strands of the patient’s DNA (shown in green) has been replaced with a “mimic” strand of DNA – an induced heteroduplex generator (IHG). This stretches the conformation of the patient’s DNA in the region where the genetic defect is located. The 4 bases used to induce heteroduplexes in the helix are shown in the middle with individual atoms multicoloured.
Using IHGs for genetic diagnosis
The effect of stretching the patient’s DNA is to change its structural conformation, which allows accurate identification of:
• Normal individuals unaffected by the disease • Those who are carriers of the disease, but who themselves are unaffected • Those who have inherited, and will develop, the genetic disease.
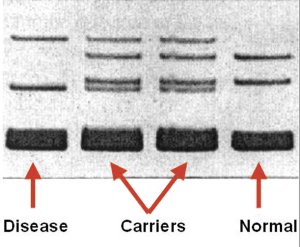
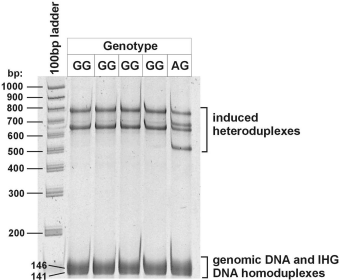
The picture shows the results of IHG diagnosis of a common blood clotting disorder (Factor V
(Above) Polyacrylamide gel electrphoresis (PAGE) readout for Factor V Leiden genotyping using IHG.
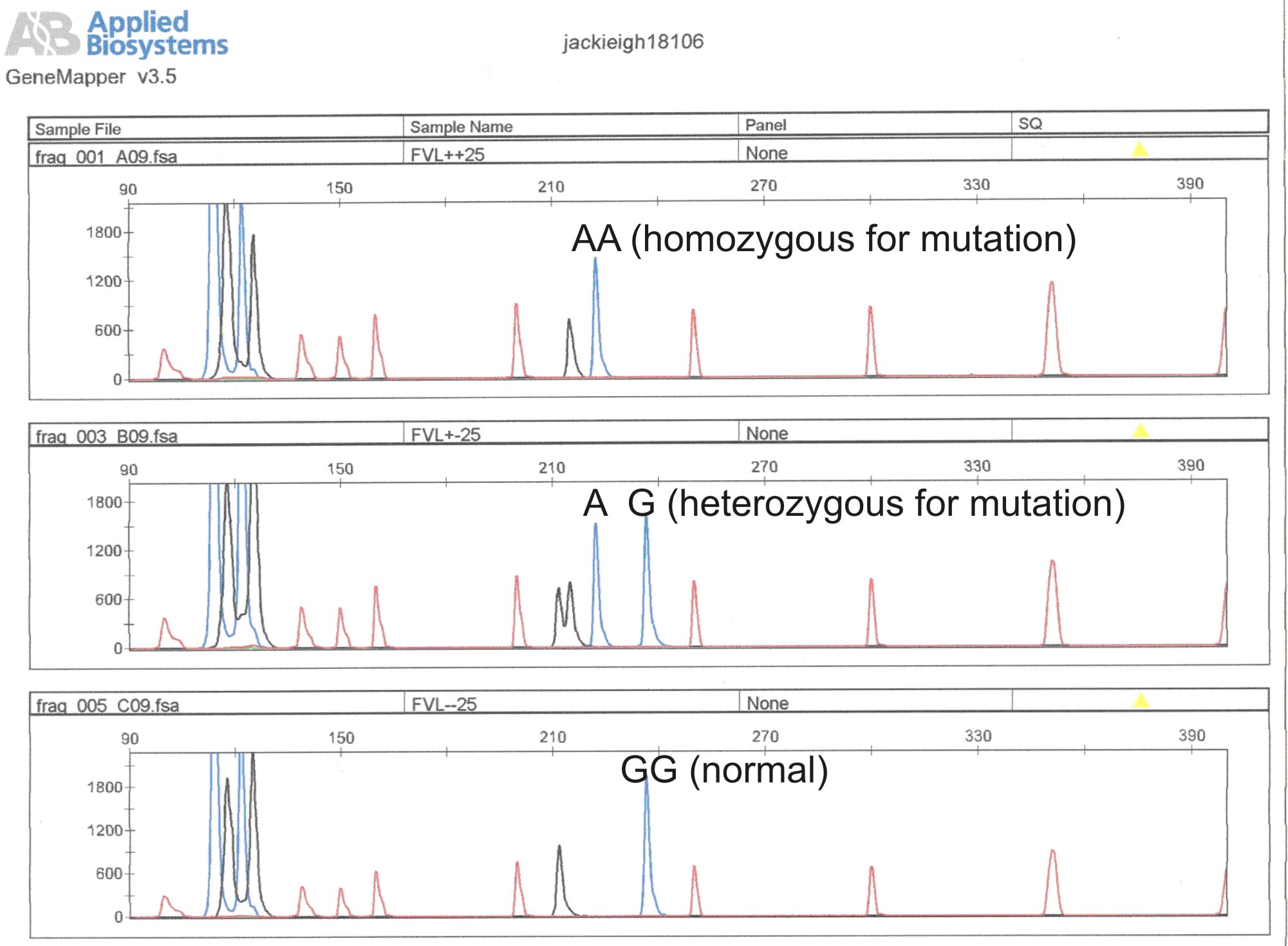
(Above) Capillary electrophoresis readout for Factor V Leiden genotyping using IHG, using an  Single nucleotide polymorphisms (SNPs) and mutations• Single nucleotide polymorphisms (SNPs) are the most frequent type of genetic variation in the human genome. They account for more than 90% of all differences between individuals. It is likely that these variable patterns of SNPs will account for many of the complex phenotypic characteristics found in humans. SNP analysis has the potential to predict susceptibility to a variety of clinical conditions including cancer, cardiovascular disease and mental illness and in making targeted drug therapy a reality. Millions of SNPs have already been identified within the human genome. SNPs are considered to be important genetic markers. This is due to their abundance within the human genome and association they may have with many genetic traits and disease susceptibility. SNPs may be situated close together within a region of DNA and can be inherited in different combinations or haplotypes. • Mutations are a class of SNP which create aberrant proteins or differences in protein expression associated with classical inherited metabolic or other diseases. Examples include: Cystic Fibrosis, Phenylketonuria, Sickle Cell Disease and Von Willebrand’s Disease.
|
||||
How is IHG technology relevant to SNP and mutation detection?Unlike many currently used DNA diagnostics, a single Induced Heteroduplex Generator (IHG) is capable of identifying single or multiple SNPs, and haplotypes. This is possible because of the unique design of the IHG. For many applications, this permits a substantial reduction in the number of tests required. Haplotyping is a great advantage because it is increasingly apparent that haplotypes, not genotypes, may be more relevant in disease and protein expression studies. IHG-based haplotyping results provide unequivocal evidence of the physical linkage of SNPs. How does the IHG reagent work?An IHG reagent mimics regions of DNA within the genes but differs from them due to the possession of one or more ‘identifiers’. These identifiers are found adjacent to the SNPs within the DNA. When human DNA (containing the SNPs in question) is mixed with IHG reagent (containing the identifiers) under certain experimental conditions, it undergoes a conformational change -the DNA bends and becomes more rigid- in a manner which is specific to the SNP or mutation causing the disease. A single IHG reagent possessing a combination of identifiers can cause unique conformational changes to occur for single or clustered SNPs and haplotypes. These changes are induced by heteroduplex formation and can be detected on various instrument platforms.
Examples: clinical PAGE minigels
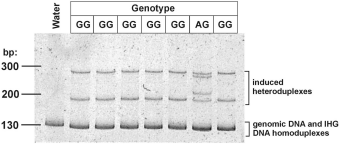
Factor V
Prothrombin PAGE minigel showing induced heteroduplexes
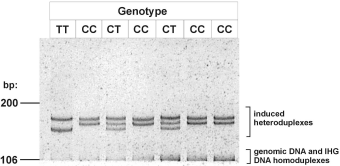
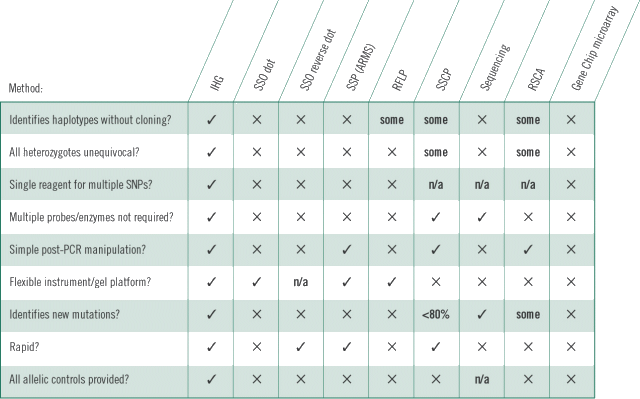
MTHFR PAGE minigel showing induced heteroduplexes What are the advantages of IHG technology?1. Simplicity: IHG-based genotyping is a simple, straightforward technique: single-tube PCR for both genomic and IHG DNAs, followed by a short mix-heat-cool step. Forget restriction enzymes, probes, hybridisation or washing steps, multiple primer pairs. 2. Ability to use multi-platform instrumentation: Results can be analysed by capillary electrophoresis (CE), or by manual or automated minigel PAGE, midigel PAGE, or maxigel PAGE, with or without fluorescently labeled primers. 3. 100% accuracy: precise detection of homozygosity and heterozygosity: no worries about allele drop-out, cross-reactivity, primer competition. 4. Identification of multiple SNPs by a single IHG reagent 5. Availability of control DNAs for all known alleles: Forget searching and bargaining for rare allele controls. All known alleles are included in the kit for use as controls. 6. Identification of haplotypes 7. Identification of new alleles: Previously unrecorded heteroduplex banding patterns indicate novel alleles. How IHG technology compares
Current methods for the detection of SNPs include the following: 1. SSO (Sequence Specific Oligonucleotide) Dot 2. SSO (Sequence Specific Oligonucleotide) Reverse Dot 3. SSP (ARMS): Sequence-Specific Primers 4. RFLP (Restriction Fragment Length Polymorphism) 5. SSCP (Single 7. Reference 8. Gene Chip Microarray/Microarray Sequencing The above methods are often intensive, expensive, time-consuming and imprecise. They are also frequently dictated by the availability of existing equipment platforms. More fundamentally extant tests are unable to determine what haplotypes an individual possesses (see above). How can a single IHG reagent identify 2 or more individual SNPs and haplotypes?
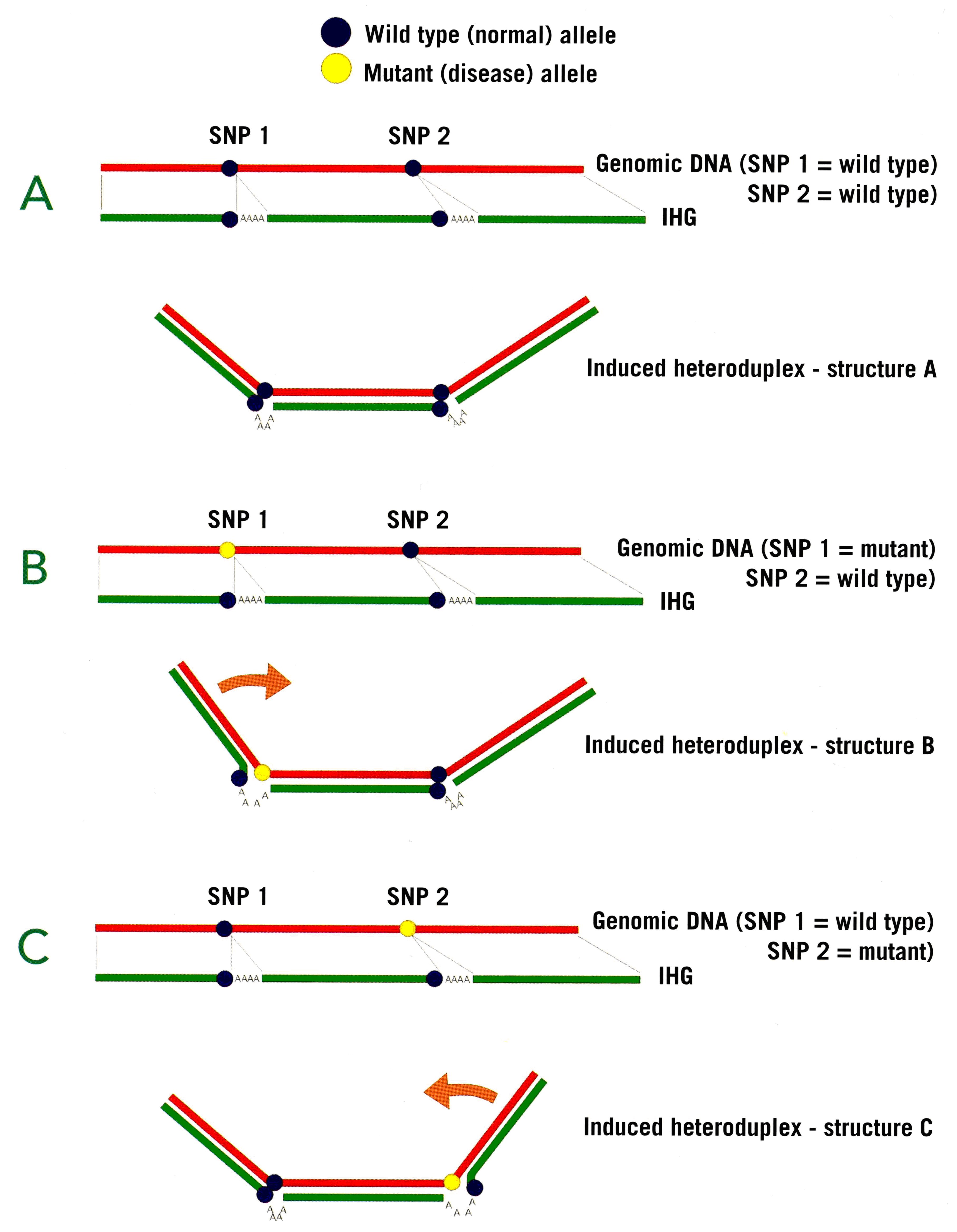
In the example above, a single IHG can detect 2 SNPs present within the same amplified region of DNA (shown in red). The wild type (normal) sequence at each of the SNPs is shown in blue, the mutant (disease) sequence is shown in yellow (see panels B and C). The 3-dimensional conformation of the heteroduplexes formed with the IHG differ (shown by different angles of the “arms”) according to whether a wild type or mutant sequence is present at one or other SNP sites.
These differences in conformations can be detected by, for example, polyacrylamide gel or minigel electrophoresis (see panel above). In the left hand column, the individual has two normal chromosomes (conformation A), which gives rise to two characteristic (arrowed) bands as shown. In the middle column, the individual has one normal chromosome and one mutated at SNP 1 (patterns A and B respectively). In the right hand column, the individual has one normal chromosome and one mutated at SNP 2 (patterns A and C respectively). In practice, up to 6 or more mutations can be detected in a single region of DNA with a single IHG reagent, by extension of the principle to include, for example, more poly(A) insertions at appropriate places in the IHG. IHGs can also detect different combinations (haplotypes) of SNPs (not shown). For example, a different pattern of bands would be observed if the individual had mutations at both SNP 1 and SNP 2 sites. Using IHGs for haplotypingMultiple single nucleotide polymorphisms (SNPs) constituting polymorphic haplotypes are close enough together to permit haplotyping with a single IHG reagent. The principle is identical to that for multiple SNP genotyping. Several IHG reagents have been developed which are capable of haplotyping. The advantage of IHG reagents over individual SNP genotyping tests is very considerable: Because IHGs characterise conformational polymorphisms over large distances, they can unambiguously identify physical linkage between SNPs, without recourse to statistical prediction of haplotypic phase. This is very important in cases where numerous haplotypes exist, giving rise to multiple ambiguities when attempting to determine haplotypes in heterozygous individuals.
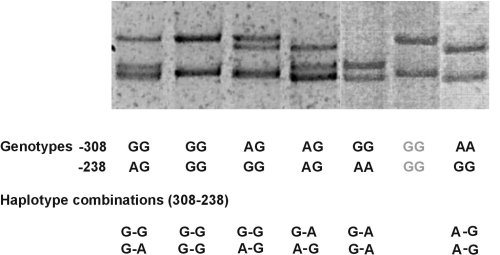
(Above) Example 1. Tumour necrosis factor a (TNF; TNFSF2) SNPs -308 G>A (rs1800629) and -238 G>A (rs361525) (3 haplotypes)
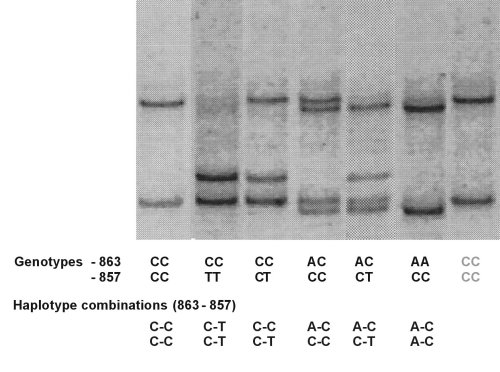
(Above) Example 2. Tumour necrosis factor a (TNF; TNFSF2) SNPs -863 C>A (rs1800630) |
||||